Section: New Results
Automatic recognition of fungi phenotype by extraction and classification of morphometric parameters
Participants : Sarah Laroui, Eric Debreuve, Xavier Descombes.
This work is made in collaboration with Aurelia Vernay (Bayer) as part of a contract with Bayer.
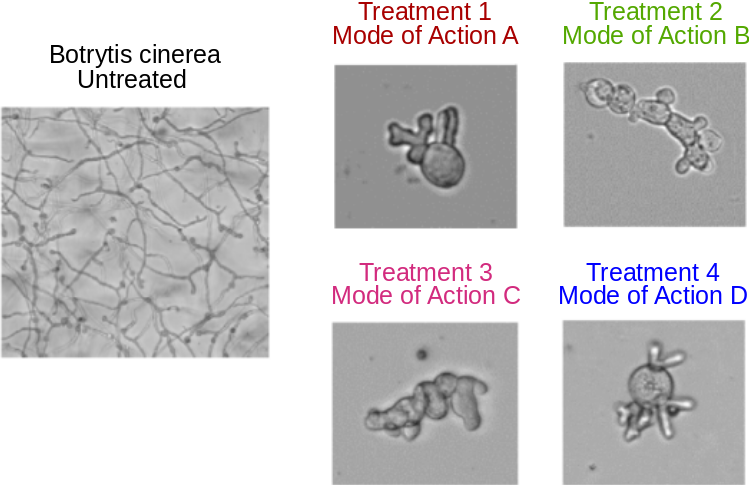
Botrytis cinerea is a reference model of filamentous phytopathogen fungi. Some chemical treatments can lead to characteristic morphological changes, or phenotypic signatures, observable with transmitted light microscopy (see Fig. 7), which could be associated with the molecule Mode of Action.
|
In this context, we developed a robust image analysis and classification method relying on morphometric characteristics to automatically detect fungi observed using transmitted light microscopy, and classify them into predefined phenotypes. The detection task has been implemented in a classical way using a combination of mathematical morphology operations and active contours. The classification task has been solved in a supervised learning context.
Since a fungus can be described as tubular extensions connected to a spore (a roundish “root” cell), we proposed to describe such an object by its skeleton together with the distances from the skeleton to the fungus boundary. The skeleton was then converted into a valued graph. We selected a dozen topological and morphological features such as the number of nodes, the length of the longest branch, or the average and variance of the per-branch average skeleton-to-boundary distances.
These features were used in a supervised machine learning framework. Specifically, a cascade of two classifiers was proposed, the first one based on a decision tree to reject non relevant phenotypes (spores and mycelium), the second one to actually determine the phenotypes of the fungi. This second classifier was a Random Forest learned on the provided learning set composed of sample fungi from two phenotypes. Note that the classification accuracy can be computed either in a per-fungus way, or in a per-image way. Indeed, a given image corresponds to a unique chemical treatment so that all the fungi it contains exhibit the same phenotype (up to the natural biological variations), which can therefore be associated to the image itself. This per-image phenotype can be obtained by a majority vote among the individual fungus phenotypes. It represents the answer the biologists need. For the 2-phenotype problem we worked on, we obtained an image classification accuracy of around 90%, which is more than encouraging. In order to allow for a future, deeper analysis of the features characterizing each phenotype, we also computed the influence of each feature on the classification accuracy.